This is an R library for estimating which individuals would respond best to a treatment on an outcome, given their values in gene transcription data.
——
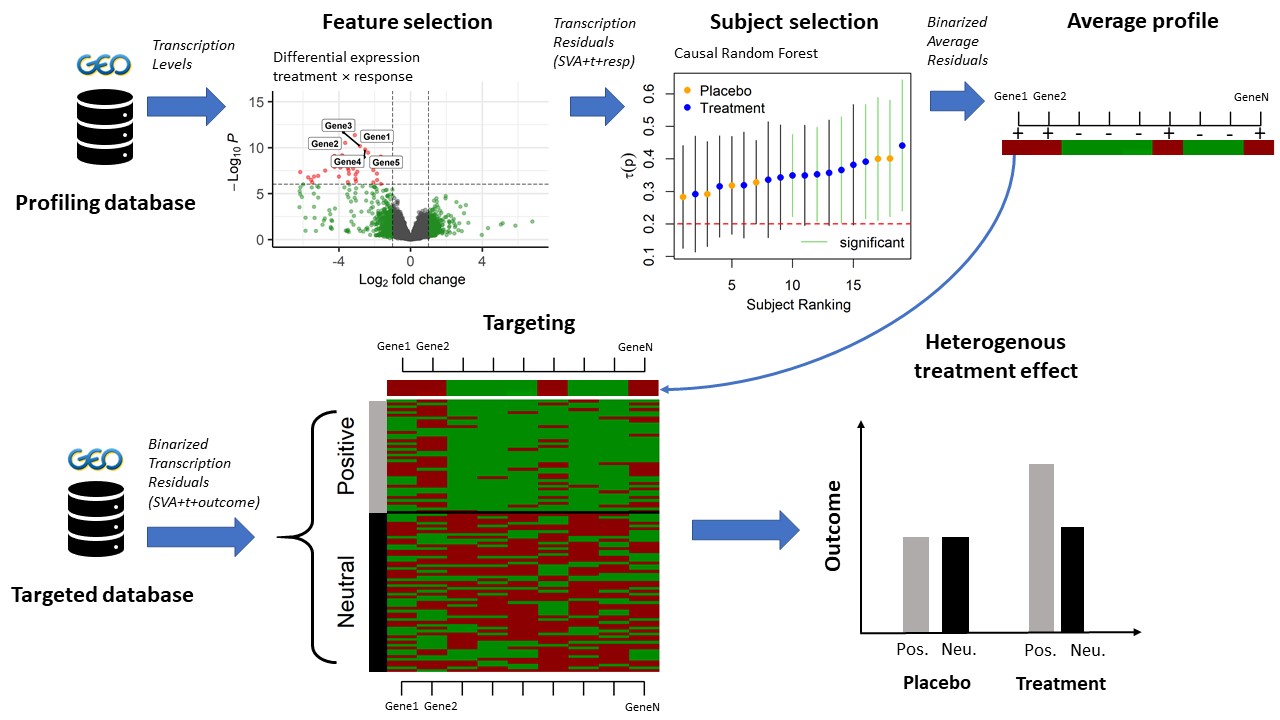
What it does
teff is a software package to estimate the expected effect of treating a single individual given the individual’s profile in some high dimensional feature data. The individual treatment effect is the expected difference in outcome between two treatments (treatment/no-treatment, drug/placebo) when feature data are kept constant. The package focuses on transcriptomic features for which surrogate covariates need to be estimated. The estimation of individual treatment effects is based on causal inference using causal random forest (CRF) as implemented in the package grf by Tibshirani et al.
The analysis strategy is based on the selection of features, subjects and extraction of a binary profile to positive, or even negative, effects of treatment. Selection of features are obtained by selecting transcription levels whose association with treatment response is significantly modulated by the treatment. Selection of individuals are those with significant treatment effects, as estimated by CRF. An average binarized profile of significant individuals is then constructed. If such profile can be identified then new individuals can be targeted and classified into groups of positive (or negative) and neutral treatment effects. When treatment and treatment response data are also available for these new individuals, the package can test whether the association between the treatment and outcome is indeed different across groups of associated positive and negative treatment effects.
Applications include:
Forecasting treatment effect of brodalumab treatment of psoriasis from gene expression data at baseline. See application.
Extracting gene expression profiles for which men and women have high differences on immune cell count in blood. In this application sex is considered as a treatment. See application.
Determining exposure environmenents with high sexual dimorphisms. Exposome Data Challenge 2021 wiinner application application.
How it does it
Feature selection is performed with transcriptome-wide differential gene expression for the interaction between treatment and response.
Individual targeting into groups of different treatment effects is based on the application of CRF to identify the individuals with significant treatment effects. Individuals with significant treatment effects are those whose confidence intervals for the treatment effect estimate do not overlap 0 (or a defined threshold).
When possible, single consensus profiles of individuals with positive, and negative, treatment effects are obtained from majority votes of features adjusted for covariates.
Additional functions
The package includes functions to extract feature and treatment data from transcriptomic studies.
Install
Install with
library(devtools) install_github(“teff-package/teff”)
You can also download the source code
- Source: teff_0.1.0.tar.gz
or binaries
- Debian Linux, R-devel, GCC: teff_0.1.0_R_x86_64-pc-linux-gnu.tar.gz
- Windows Server 2022, R-devel, 64 bit: teff_0.1.0.zip
- macOS 10.13.6 High Sierra, R-release, CRAN’s setup: teff_0.1.0.tgz
and, for instance, install for Windows with
library(devtools) install_local(“teff_0.1.0.zip”, dependencies=TRUE)
See application to Brodalumab treatment of psoriasis
Cite: Alejandro Caceres and Juan R Gonzalez. teff: estimation of Treatment EFFects on transcriptomic data with casual random forest, 2022; under submission.